Pedro Almanda
Pedro M. Pereira
Siân Culley
Ghislaine Caillol
Fanny Boroni-Rueda
Christina L. Dix
Guillaume Charras
Buzz Baum
Romain F. Laine
Christophe Leterrier
Ricardo Henriques
In this paper, Henriques lab proposes an open source, cheap and well documented hardware system to automate on microscope the treatment and labelling of samples. It makes event base microscopy and live to fix super-resolution imaging easier and more reproducible, thus opening interesting venues for smart microscopy.
Publication:
Automating multimodal microscopy with NanoJFluidics, Nature Communications volume 10, Article number: 1223 (2019)
#
SuperResolution #SmartMicroscopy #DIY #LegoBased #EventBasedMicroscopy #DNApaint #OpenSource #MicroscopyTechnique

A blog post by Adrien Méry
After my engineering degree, I spent my PhD exploring the mechanical response of tissues using optogenetics and tissues engineering. This was also when I developed a passion for science communication and started working as a freelance scientific copywriter. Taking part to the Idylle blog is thus the perfect opportunity to combine both my love for pop-sci and cool research tools.
.png)
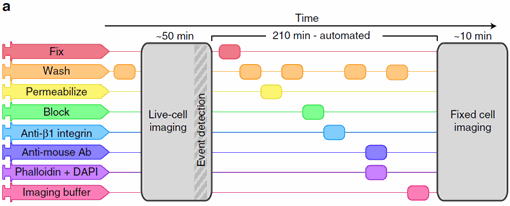
Figure 1: An imaging protocol including automated fix and labelling of the cells. From Almanda 2019 (10.1038/s41467-019-09231-9).
Nano-J fluidics : an easy gateway to smart microscopy?
With the microscopes and especially the computer power available in biology labs nowadays, complex experiments based on event detection are conceivable. The missing link however is an affordable hardware system to react to the detection of event that does not depend on the experimenter. Nano-J fluidics fills this gap and allows the automation of microscopy experiment and frees up time for the experimenter.
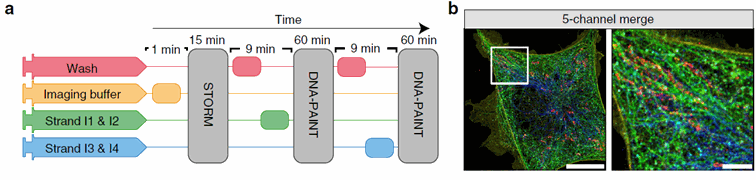
Figure 2: The Nano-J fluidics system allows to easily multiplex STORM and DNA paint imaging without having to move the sample sample. On the left, we see a time-based representation of the protocol using 4 different pumps. On the right, the final merged image obtained. Taken from Almanda 2019 (10.1038/s41467-019-09231-9)
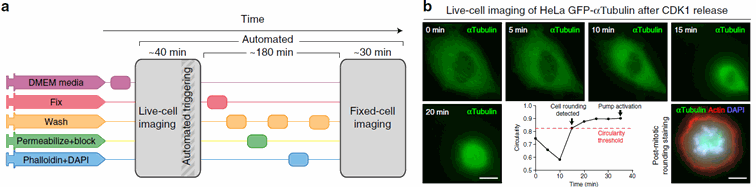
Figure 3: The system also allows event based microscopy : here for instance mitosis imaging. Once the rounding of the cells is detected, the fixation and labelling process is triggered, and a new round of imaging follows. Taken from Almanda 2019 (10.1038/s41467-019-09231-9)
A Lego syringe pump system, what for?
All the interest of the technique resides in the fact that you can change the medium of your cells with just a click on your computer. This allows to perform complex experiments that would be tedious for the user.
You can use this system to simply perform live to fixed cells experiments or multiplexing DNA paints imaging without moving your sample. In this last case, Nano-J fluidics allows a quick change of the fluorescent DNA strands while keeping the cells in view. This also comes handy for super resolution imaging.
To me however, the main interest of this system is the possibility to make event-based microscopy. The team here gives the simple example of mitosis imaging: when the rounding of the cells is detected, the fixation protocol is triggered and the cells are fixed in their status. But more specific experiments, based on the detection of other cells actions can be imagined.
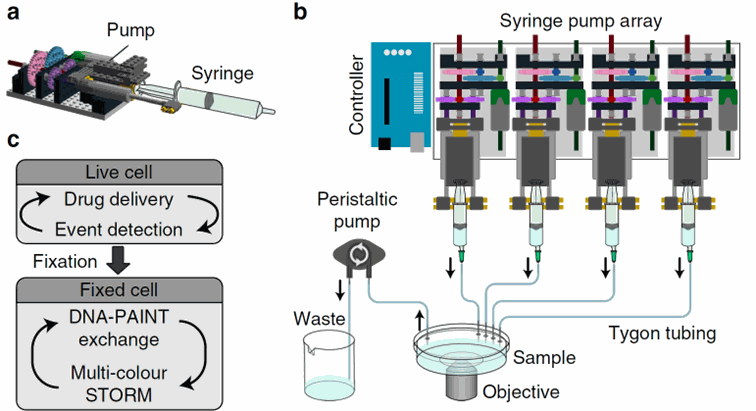
Figure 4: Schematic of the pump system. Each pump is assembled with a syringe and a pump based on Lego bricks. Several pumps can be assembled and are controlled by an Arduino system connected to a computer. Taken from Almanda 2019 (10.1038/s41467-019-09231-9)
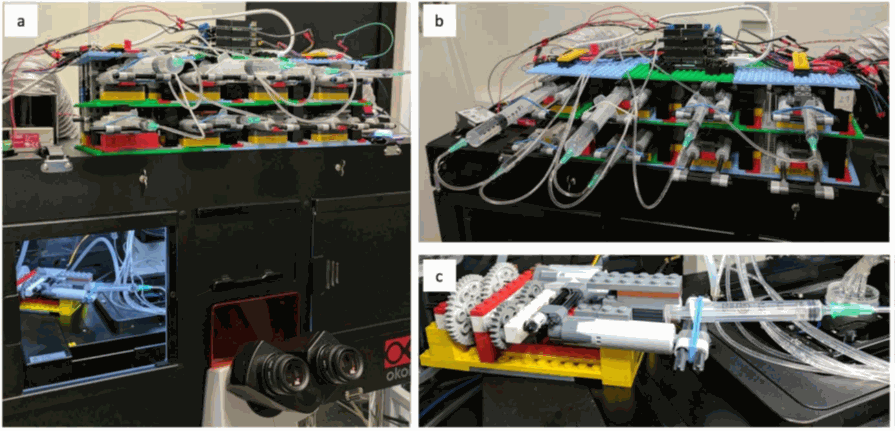
Figure 5: A pump array of 8 pumps easily fits in the microscopy room and a pump can even
be installed
in the incubator to keep the medium at 37°C. Taken from Almanda 2019 (10.1038/s41467-019-09231-9)
A very simple system
The huge interest of this system is its cost. The equipment is finally very cheap and easily assembled following the steps given online on the Gitlab of the team. It is composed of easily found components: Arduino microcontrollers, tubing, syringes, and … Lego bricks for the pumps!
Once assembled and calibrated, the system is piloted using a Fiji extension. An interface allows the user to program the control of the pump, based on a timed protocol (to perform a sequence of imaging using different probes) or the detection of events. Here, the team takes the example of the rounding of the cells, but we can imagine different cues detected with the microscope (fluorescent change, cell movement or blebbing…)
References:
Schnitzbauer, J., Strauss, M., Schlichthaerle, T. et al. Super-resolution microscopy with DNA-PAINT. Nat Protoc 12, 1198–1228 (2017). https://doi.org/10.1038/nprot.2017.024
Tam J, Cordier GA, Borbely JS, Sandoval Álvarez Á, Lakadamyali M (2014) Cross-Talk-Free Multi-Color STORM Imaging Using a Single Fluorophore. PLOS ONE 9(7): e101772. https://doi.org/10.1371/journal.pone.0101772
Liu, Z., Lavis, L. D., & Betzig, E. (2015). Imaging Live-Cell Dynamics and Structure at the Single-Molecule Level. In Molecular Cell (Vol. 58, Issue 4, pp. 644–659). Cell Press. https://doi.org/10.1016/j.molcel.2015.02.033
MORE LIFE SCIENCE TOOLS TO FOLLOW
You want the latest on life science tools to follow, test and more?